
A peer-reviewed electronic journal.
Copyright is retained by the first or sole author, who grants right of first publication to the Practical Assessment, Research & Evaluation.
Permission is granted to distribute this article for nonprofit, educational purposes if it is copied in its entirety and the journal is credited.
PARE has the right to authorize third party reproduction of this article in print, electronic and database forms.
Volume 19, Number 12, August 2014 ISSN 1531-7714
Sample Size Determination for Regression Models
Using Monte Carlo Methods in R
A. Alexander Beaujean, Baylor University
A common question asked by researchers using regression models is, What sample size is needed for
my study? While there are formulae to estimate sample sizes, their assumptions are often not met in
the collected data. A more realistic approach to sample size determination requires more
information such as the model of interest, strength of the relations among the variables, and any
data quirks (e.g., missing data, variable distributions, variable reliability). Such information can only
be incorporated into sample size determination methods that use Monte Carlo (MC) methods. The
purpose of this article is to demonstrate how to use a MC study to decide on sample size for a
regression analysis using both power and parameter accuracy perspectives. Using multiple regression
examples with and without data quirks, I demonstrate the MC analyses with the R statistical
programming language.
A question posed in the design of many research
studies is: What sample size is needed? Being able to
answer this question is important because institutional
research boards and most granting agencies require that
investigators specify the size of the sample they intend
to collect. In addition, most reporting guidelines for
education, psychology, and health professions research
require authors to state how they determined their
sample size (e.g., American Educational Research
Association, 2006; American Psychological Association
Publications and Communications Board Working
Group on Journal Article Reporting Standards, 2008;
Moher et al., 2010). More practically, conducting a
study with the wrong sample size can be costly–having
too few participants results in the inability to find
effects or precisely estimate their values, while having
too many participants results in wasting the
investigators’ valuable resources.
Typically investigators determine the needed
sample size via some table or formulae in a textbook
(e.g., Murphy & Myors, 1998), or by using specifically-
designed software (e.g., Faul, Erdfelder, Lang, &
Buchner, 2007). While this approach can be useful for
simple projects, the assumptions used in these
calculations often do not hold in the actual data. An
alternative approach to determining the required
sample size is to use a Monte Carlo (MC) study.
MC studies use random sampling techniques,
typically done through computer simulation, to build
data distributions (Beasley & Rodgers, 2012).
Researchers often use them as an empirical alternative
to solve problems that are too difficult to solve through
statistical or mathematical theory (Fan, 2012). There are
a variety of uses for MC studies, ranging from
understanding statistics with unknown sampling
distributions to evaluating the performance of a
statistical technique with data that do not meet the
technique’s assumptions.
Previously, Muthén and Muthén (2002) showed
how MC methods can be useful to determine sample
size for a structural equation model (SEM). While this
approach has been praised (Barrett, 2007), it requires
using specialized proprietary software and has not been
readily accessible to a wide audience. Further, while
regression is a specific type of
SEM (Hoyle & Smith, 1994), scholars who rely on
regression analysis might not understand how they are
Practical Assessment, Research & Evaluation, Vol 19, No 12 Page 2
Beaujean, Monte Carlo Sample Size Determination
related. Thus, they may have ignored the MC approach
to sample size determination. Consequently, there is a
need to show how to use MC methods, using freely
accessible software, to determine the needed sample
size for use with regression models.
The purpose of this article is to demonstrate the
use of a MC study to determine the required sample
size for a multiple regression analysis. I demonstrate
such analyses using the R (R Development Core Team,
2014) statistical programing language, which is open
source, available for multiple operating systems, has
extensive data simulation facilities, and has great
flexibility that is unmatched by most other statistics
programs (Kelley, Lai, & Wu, 2008).
Power Analysis
Power analysis was developed concurrently with
null hypothesis significance testing (NHST), although it
wasn’t until Jacob Cohen’s work in the 1960s that it
became popular (Descôteaux, 2007). NHST pits two
competing hypotheses against each other: the null (H
0
)
and the alternative (H
a
). When used for power analysis,
H
0
is usually specified to be that the parameter of
interest equals zero, while H
a
is specified to be that the
parameter does not equal zero. The needed sample size
in this scenario refers to the number of observations
required to reject H
0
.
Power analysis involves four interrelated concepts:
a) sample size;
b) type 1 error (α);
c) type 2 error (β) or statistical power (1–β); and
d) effect size (Cohen, 1988).
The concepts are deterministically related to each other,
meaning that if three are known, so is the fourth. Thus,
providing values for type 1 error, type 2 error (or
power), and the effect size will provide the needed
sample size.
Type 1 and type 2 error values are relatively
straightforward to provide, but an effect size (ES) is
more difficult to specify (Cohen, 1992). Not only are
there different types of ESs that use different metrics
and are only useful with certain kinds of data (Grissom
& Kim, 2005), but there is usually little knowledge of
what constitutes a typical or clinically-relevant ES
magnitude for a given field of study (Hill, Bloom,
Black, & Lipsey, 2008). In regression, the ES measure
is usually a regression coefficient or the amount of the
variance the model explains of the outcome variable
(i.e., R
2
).
Parameter Accuracy
Many scholars have sharply criticized NHST over
the last two decades (Cumming, 2014; Wilkinson &
American Psychological Association Science
Directorate Task Force on Statistical Inference, 1999).
More recently, scholars have begun placing the NHST-
related power analysis procedure under scrutiny as well
(Bacchetti, 2010, 2013). As an alternative to
determining sample size through a power analysis is to
determine it using accuracy in parameter estimation
(AIPE; Kelley & Maxwell, 2003; Maxwell, Kelley, &
Rausch, 2007). Although the two approaches are not
mutually exclusive (Goodman & Berlin, 1994), their
philosophies are very different. In the power analysis
perspective, interest lies in having just enough accuracy
so that the value of a parameter estimate is statistically
different that zero (i.e., rejecting H
0
). In the AIPE
perspective, interest lies in the accuracy of a
parameter’s estimate, no matter if the estimate’s value is
zero or any other variable. Kelley and Maxwell (2003)
argued that the AIPE approach leads to a better
understanding of an effect than the power approach.
As NHST is embedded in the power approach, the
only new knowledge it provides is whether a parameter
is different than zero. Obtaining sufficiently accurate
parameter estimates, however, can lead to knowledge
about the parameter’s likely value.
The accuracy component in AIPE is defined as the
discrepancy between a parameter’s estimated value and
its true value in the population (Hellmann & Fowler,
1999). It is measured by the mean square error of a
parameter’s estimator, which is comprised of two
additive parts. The first is variance, the inverse of
which is precision. The second part is bias. Thus, when
a parameter estimator is unbiased, accuracy and
precision are directly related to each other.
The square root of a parameter’s variance is its
standard error, which is used for creating a confidence
interval (CI; Cumming & Finch, 2005). Consequently,
one way to assess the accuracy of a parameter estimate
is by examining the width, or half-width, of its CI. The
half-width is the halved difference between the upper-
bound and lower bound of the CI. The narrower the
CI (i.e., the smaller the half-width), the more precise
the parameter estimate and more certainty there is that

Practical Assessment, Research & Evaluation, Vol 19, No 12 Page 3
Beaujean, Monte Carlo Sample Size Determination
the observed parameter estimate closely approximates
the corresponding population value.
Traditional Methods for Estimating Power and
Parameter Accuracy
Many have written about the methods involved in
determining sample size for a regression analysis using
traditional power analysis (Dupont & Plummer, 1998;
Maxwell et al., 2007). It involves the following steps: (a)
review similar studies to find their ES values; (b)
determine the expected ES values for the current study;
(c) set α and power at the desired values; and (d)
calculate the sample size needed to find the expected
effect is statistically significant at the givenα level
while retaining the desired amount of power (Cohen,
1992). This calculation can be done analytically or
through computer programs designed for such analyses
(for a list, see Kelley & Maxwell, 2012, p. 199).
Determining sample size for a regression using the
AIPE perspective involves a similar set of steps: (a)
determine the predictor and outcome variables; (b)
review studies that used similar variables and find the
values of the relations between the predictor and
outcome variables as well as the relations among the
predictor variables; (c) determine the expected variable
relations for the current study; (d) set the desired half-
width of the CI and confidence level (e.g., 95%, 90%);
and (e) calculate the sample size needed to find the
desired CI half-width for a given confidence level and
set of variable relations. This calculation can be done
manually or via a computer program (Kelley, 2007;
Kelley & Maxwell, 2003).
Using either the power- or AIPE-based formulae
and procedures to determine sample size can be useful
for very simple situations, but has problems when it
comes to more practical research situations (Bacchetti,
2013). For example, they typically assume there are no
missing data and that the collected data will meet the
assumptions for the statistical tests of interest—
assumptions that are often not met. An alternative to
the traditional formulae-based method is to use a MC
study, which can estimate not only the required sample
size from both the power and AIPE perspectives but
also can incorporate data quirks such as missing values
and assumption violations.
Monte Carlo Methods for Determining
Sample Size
Muthén and Muthén (2002) showed how MC
methods can be useful for determining the sample size
needed for SEMs based on a power analysis. Generally,
the procedure they outlined requires simulating a large
number (m) of samples, each of size n, from a
population with hypothesized parameter values. The
model of interest (e.g., regression) is then estimated for
each of the m samples and the set of m parameter
values and standard errors are then averaged. The
required sample size is the smallest value of n that
produces the desired power for the parameters of
interest contingent on the simulated data meeting
certain quality criteria, which I discuss in the
subsequent section. Muthén and Muthén did not
discuss parameter accuracy, but this can easily be
incorporated by select the sample size based on the
data having the desired CI half-width instead of having
the desired power.
Criteria to Determine Monte Carlo Study’s Quality
.
The following statistics can be useful to determine
the quality of the simulated data in a MC study: (a)
relative parameter estimate bias, (b) relative standard
error bias, and (c) coverage. Relative parameter estimate
bias is:
(1)
where
is the hypothesized (pre-set) value of the
parameter, and
is the average parameter estimate
from the m simulated samples. Relative standard error bias
is:
(2)
where
is the standard deviation of the m
parameter estimates, and
is the average of the m
estimated standard errors for the parameter. Coverage is
the percent of the m simulated samples for which the
(1–α)% CI contains θ. Table 1 contains Muthén and
Muthén’s (2002) suggested criteria for these statistics.
Once they are met, power is calculated as the proportion
of the m simulated samples for which H
0
(i.e., θ = 0)
is rejected using the specified α level.

Practical Assessment, Research & Evaluation, Vol 19, No 12 Page 4
Beaujean, Monte Carlo Sample Size Determination
Table 1.
Criteria for Monte Carlo Data Quality
Statistic Criteria
Coverage Between .91 and .98
Relative
parameter bias
Absolute value ≤ .10 for all
model parameters
Relative standard
error bias
Absolute value ≤ .10 for all
model parameters
Absolute value ≤ .05 for the
parameters of major interest
Note. Taken from Muthén & Muthén (2002, pp.
605-606).
Decisions to Make in a Monte Carlo Sample Size
Study.
Figure 1 contains the required steps for using a
MC study to determine the needed sample size for a
regression analysis. Before simulating the data (Step 5),
a number of decisions need to be made. First,
determine the regression model to study, which
includes all pertinent predictor variables as well as the
nature of the associations between the predictor
variables and outcome variable. Creating a detailed path
diagram can greatly facilitate this step (Boker &
McArdle, 2005).
Second, decide on population values for every
parameter. This includes the regression coefficients, the
scale and reliability of all variables, the amount of
residual variance, and the covariances among the
predictor variables. This step will typically be easier if
the variables are standardized, as this makes the
covariances become correlations, the regression
coefficients become standardized, and the intercept
become zero. After determining the parameter values,
it is important to check that the implied covariance
matrix values (i.e., the covariances based on the
population parameter values) are as expected. As with
the first step, path diagrams can be very helpful here as
well.
Inherent in the second step is the decision on the
ES value. That is, by specifying the values for the
1.
Decide on regression model.
1.1. Draw a path diagram of the model to account for all intended relationships (optional).
2 Decide on population values for all parameters in model, including: regression coefficients,
scale and reliability of the variables, the amount of residual variance (
), and covariance
among the predictor variables. Standardizing the variables makes this step easier.
2.1. Check values of the implied covariance matrix to make sure they are as expected.
3. Decide on any data quirks, such as missing values or assumption violations (optional).
4. Decide on the technical aspects of the MC simulations:
4.1. Type 1 error rate (
α
), which also determines the CI.
4.2. Desired power (1–
β
) or confidence interval half-width.
4.3. Number of samples to simulate (m).
4.4. Sample size (n) or range of sample sizes.
4.5. Random seeds (at least two).
5. Simulate the m samples of the regression model from Step 2.
6. In the simulated data, check (cf. Table 1):
6.1. Relative parameter and standard error biases.
6.2. Coverage.
7. If the values in Step 6 are acceptable, examine the power or parameter accuracy of the
parameters of interest. If the values are not high enough, increase n and repeat Steps 5 and 6.
8. Repeat Steps 5 - 7 using a different random seed.
9. Compare results of simulated data from both random seeds.
9.1. If they converge, no need for further simulations of current scenario.
9.2. If they do not converge, repeat Steps 5 - 8 using different random seeds or larger values
of m.
Figure 1. Steps for sample size planning of a regression analysis using a Monte Carlo study.

Practical Assessment, Research & Evaluation, Vol 19, No 12 Page 5
Beaujean, Monte Carlo Sample Size Determination
regression coefficients and the predictor variables’
covariance, R
2
(or any other regression ES measure) is
already determined. I elaborate on these relations more
in the first example.
The third step is to decide on any data quirks, such
as having missing values or violating any assumptions.
This step is optional as its usefulness depends on the
variables and population from which the data will be
collected. The fourth step requires decisions about
values for the technical aspects of the MC simulations.
This includes: α, power, the number of samples to
simulate (m), the sample sizes (n), and the random
seeds. The value for m should be large, as the goal is to
produce stable results and large values for m tend to
produce quality simulations. Muthén and Muthén
(2002) suggested setting m to 10,000, but this may be
excessive for simpler regression models with no data
quirks (Skrondal, 2000). The initial n to use is
somewhat arbitrary. If there is no reason to select one
specific value, then it might be better to decrease m and
simulate samples for a sequence of ns.
The random seed is an integer used to initialize the
pseudo-random number generation for the simulations
(Marsaglia, 2003). A given seed value generates the
same sequence of numbers, so using the same seed
value will simulate the exact same data while using
different seed values will simulate different data. Using
different seed values is comparable to taking different
independent samples from the same population. At a
minimum, the MC study should be done at least twice
using two different, randomly selected seed values. The
results from the two different simulations should
converge—that is, they should both point towards
using roughly the same sample size. If that is the case,
then there is no need for further MC simulations of
that particular scenario. Otherwise, additional
simulations using additional seeds may be needed.
Presentation of Following Material
In what follows, I present two examples of the MC
method for determining sample size for regression
analysis using R. The first example is a typical multiple
regression model. In the second example, I extend the
first example by adding data quirks involving: (a)
missing data, (b) the outcome variable’s distribution,
and (c) variable reliability. As I discuss a given analysis,
I present R syntax to conduct the analysis in a separate
text box for one random seed. The words in gray
following a pound sign (
#
) in the syntax are comments,
so R ignores them. For those with no previous
experience using R, Venables et al., (2012) provide a
good introduction.
Example 1: Multiple Regression
Background
A typical regression power analysis involves
examining a model’s R
2
value or a change in R
2
from
one model to another.
1
The MC method requires more
information as it needs values for the relations between
the outcome variable and all the predictor variables as
well as the relations among all the predictors. Once
those are specified, then the model’s R
2
can be
calculated using Equation 3. If the p predictor variables
and single outcome variable are mean-centered, then
(3)
where
is the p × 1 column vector of
correlations between each of the predictor variables
and the outcome, b
YX
is the p × 1 column vector of
regression coefficients of each of the predictor
variables, C
XX
and V
XX
are the p × p correlation and
covariance matrices, respectively, of the predictor
variables, and
is the variance of the outcome
(Christensen, 2002). A little manipulation of Equation 3
reveals that the set of standardized regression
coefficients, b*, can be estimated by
(4)
Regression Model
Kelley and Maxwell (2003) presented an example
of a sample size study for a regression model with three
predictor variables. The predictor variables’ correlations
with each other, R
XX
, as well as the correlations
between each of the predictor variables and the
outcome,
, are:
!! "! #!
"! !! !$
#! !$ !!
%&'()
*+
$!
,!
!
%
To calculate the R
2
value, plug the values into
Equation 3; likewise, to calculate the standardized
regression coefficients, plug in the known values into
1
Cohen (1988) used the
-
value, but it is just a
transformation of
R
2

Practical Assessment, Research & Evaluation, Vol 19, No 12 Page 6
Beaujean, Monte Carlo Sample Size Determination
Equation 4. The values for the three regression
coefficients, respectively, are 0.66, 0.05, and –0.30.
# correlations between predictors and outcome
xy <- c(0.5, 0.3, 0.1)
# correlation matrix among predictors
C <- matrix(c(1, 0.4, 0.6, 0.4, 1, 0.05, 0.6, 0.05, 1),
ncol = 3)
# R2
R2 <- t(xy) %
*
% solve(C) %
*
% xy
# standardized regression coefficients
b <- solve(C) %
*
% xy
Simulating data for regression models with
multiple predictors can be tricky, as it has to account
for the relationships among all the variables. Using path
diagrams eases this process, as proper diagrams show
all the model parameters. A path diagram for Kelley
and Maxwell’s (2003) example with the parameter
values is in Figure 2.
Figure 2. Path model of multiple regression for
Example 1.
Another advantage of using path diagrams is that
they can facilitate specifying the regression model in R,
as the lavaan (Rosseel, 2012) package uses path models
for input. The lavaan operators for specifying path
models are given in Table 2. Beaujean (2014) contains
some worked examples of regression models using
lavaan.
The following syntax specifies the regression
model (i.e., Figure 2) in lavaan using the known values.
# load lavaan
library( lavaan)
# specify regression model with population values
pop.model<-'
# regression model
y ~ 0.66
*
x1 + 0.05
*
x2 + -0.30
*
x3
# predictor variable correlations
x1~~0.40
*
x2 + 0.60
*
x3
x2~~0.05
*
x3
# residual variance
y ~~ 0.6854
*
y
'
Table 2. lavaan Operators for Specifying Path
Models.
Syntax
Command Example
~
Regress onto Regress B onto A:
B ~A
~~
(Co)varaince
Variance of A:
A ~~A
Covariance of A and B:
A ~~B
~1
Constant/mean/
intercept
Regress B onto A, and
include the intercept in the
model:
B ~A
B ~1
or
B ~1 + A
=~
Define reflective
latent variable
Define Factor 1 by A-D:
F1 =~A+B+C+D
*
Label or
constrain
parameters
(the
label/constraint
has to be pre-
multiplied)
Label the regression of Z
onto X as b:
Z ~b
*
X
Make the regression
coefficient 0.30:
Z ~.30
*
X
After specifying the regression model and deciding
on the population values, Step 2.1. requires checking to
make sure the specified values produce the correct
results. This can be done in R by estimating the model
using the specified parameter values and examining the
results. To do this in lavaan, use the
sem()
function with
the
fixed.x=FALSE
argument. The
fixed.x=FALSE
argument is required when fixing a predictor variable’s
variance or covariance. The
fitted()
function returns the
model-based means and covariances (i.e., those implied
by using the fixed parameter values), while the
cov2cor()
converts a covariance matrix to a correlation
matrix. As I used standardized values for the
parameters, the covariance and correlation matrices are
identical for this example.

Practical Assessment, Research & Evaluation, Vol 19, No 12 Page 7
Beaujean, Monte Carlo Sample Size Determination
# check model parameters
pop.fit <- sem(pop.model, fixed.x = FALSE)
summary(pop.fit, standardized = TRUE, rsquare =
TRUE)
# model implied covariances
pop.cov <- fitted(pop.fit)$cov
# model implied correlations
Cov2cor(pop.cov)
The resulting model-implied correlations are the
same as the values given by Kelley and Maxwell (2003),
indicating that the regression parameters are specified
correctly for the MC simulations. As I do not have any
data quirks in this initial example, the next step is to
decide on the technical aspects of the MC simulations.
Kelley and Maxwell (2003) wrote that with n = 237, all
the 95% CI half-widths will be ≤ 0.15. Thus, I set the
following: (a) α: .05; (b) CI half-width : ≤ 0.15; (c) n :
237; (d) random seeds : 565 and 54447; and (e) m : 500.
I selected a relatively small number for m as this model
is not very complex.
Because lavaan cannot run the MC study directly, I
use the simsem package (Pornprasertmanit, Miller, &
Schoemann, 2012) for the simulations. This package is
designed for MC studies of sample size and accepts
lavaan model specification. All subsequent R syntax is
for simsem functions.
Conducting a MC study in simsem requires
specifying two models. The first generates the samples,
and is the one I previously specified. The second model
estimates parameters from the simulated samples.
Typically, the second model will be the same as the first
except it will not contain values for the parameters.
# multiple regression data analysis model
analysis.model <- ' y ~ x1 + x2 + x3
'
To simulate the data, use the
sim()
function. Its
main arguments are: (a) the number of samples, m
(
nRep
); (b) the data generating model (
generate);
(c) the
model to analyze the data (
model
); (d) the sample size
(
n
); (e) the lavaan function to use for the analysis
(
lavaanfun
); and (f) the random seed (
seed
). The
multicore
argument is optional, but if set to
TRUE
then
R will use multiple processors for the simulation. This
can considerably lessen the time required to create the
data.
# load simsem package
library(simsem)
# simulate data
analysis.237 <- sim(nRep = 500,
model=analysis.model, n = 237,
generate=pop.model, lavaanfun = "sem",
seed=565, multicore=TRUE)
The
summaryParam()
function, using the
detail=TRUE
argument, returns the averaged values of
interest from the simulated samples. Using the
alpha =
0.05
argument makes all CIs set at 95%. In Table 3, I
explain each of the output values.
Table 3
Returned values from
simsem
’s
summaryParam()
function.
Name Statistic
Estimate.Average Average parameter estimate
across all samples.
Estimate.SD Standard Deviation of parameter
estimates across all samples.
Average.SE Average of parameter standard
errors across all samples.
Power..Not.equal.0.
Power of parameter at given
α
.
a
Std.Est Average standardized parameter
estimate across all samples.
Std.Est.SD Standard deviation of
standardized parameter estimates
across all samples.
Average.Param Specified parameter value.
Average.Bias The difference between average
parameter estimate and specified
parameter value.
Coverage Coverage of parameter using
(1–
α
)% confidence intervals.
a
Rel.Bias Relative parameter bias.
Std.Bias
Standardized parameter bias
.
/
0
Rel.SE.Bias
Relative standard error bias.
Average.CI.Width
Average (1–
α
)% confidence
interval width (not half-width).
a
SD.CI.Width
Standard deviation of (1–
α
)%
confidence interval width.
a
Note. To produce all the statistics requires using the
detail=TRUE
argument.
a
α= .05 by default, but can be changed using the alpha
argument.
Practical Assessment, Research & Evaluation, Vol 19, No 12 Page 8
Beaujean, Monte Carlo Sample Size Determination
# return averaged results from simulated data
summaryParam(analysis.237, detail = TRUE, alpha
= 0.05)
The values from the MC study are in the top of
Table 4. The relative biases and coverage are within
specified values. As expected, the 95% CI half-widths
are all ≤ 0.15. Power is ≥ .80 for X1 and X3’s
regression coefficients, but for X2 it is only .12. This
illustrates the difference between the power and AIPE
approaches as parameter estimates can be accurate but
not powerful, especially when they are very close to
zero.
Unknown Sample Size
If the sample size to use is unknown, then instead
of giving a single value for the n argument give a range
of values using the sequence function,
seq()
. For
example, to examine power and accuracy for values
from n = 200 to n = 400, increasing by increments of
25, use
seq(200,400,25)
for the
n
argument. This
produces one simulation with n = 200, one with n =
225, and so forth. To increase m at each n, wrap the
seq()
function inside the replicate function,
rep()
. For
example,
rep(seq(200,400,25), 50)
repeats the 200-400
sequence 50 times (i.e., m = 50). The goal here is not to
meet the criteria in Table 1, but to hone in on plausible
values of n using smaller values of m. After finding
some possible values for n, complete the MC study
with a single sample size and a much larger m.
# simulate data with sample sizes from 200-400
increasing by 25 (m=50)
analysis.n <- sim(nRep = NULL,
model=analysis.model,
n = rep(seq(200,400,25), 50), generate=pop.model,
lavaanfun = "sem", seed=565,
multicore=TRUE)
Saving the results from the multiple sample size
simulations allows for the creation of both a power
curve and an accuracy curve, which is a graph of the
(1–α)% CI width as a function of sample size. To
create the the former, use the
plotPower()
function with
the parameter of interest as the value for the
powerParam
argument and theαvalue as the value for
the alpha argument. To crate the latter, use the
plotCIwidth()
function with the parameter of interest as
the value for the
targetParam
argument and 1–α as the
value for the
assurance
argument.
2
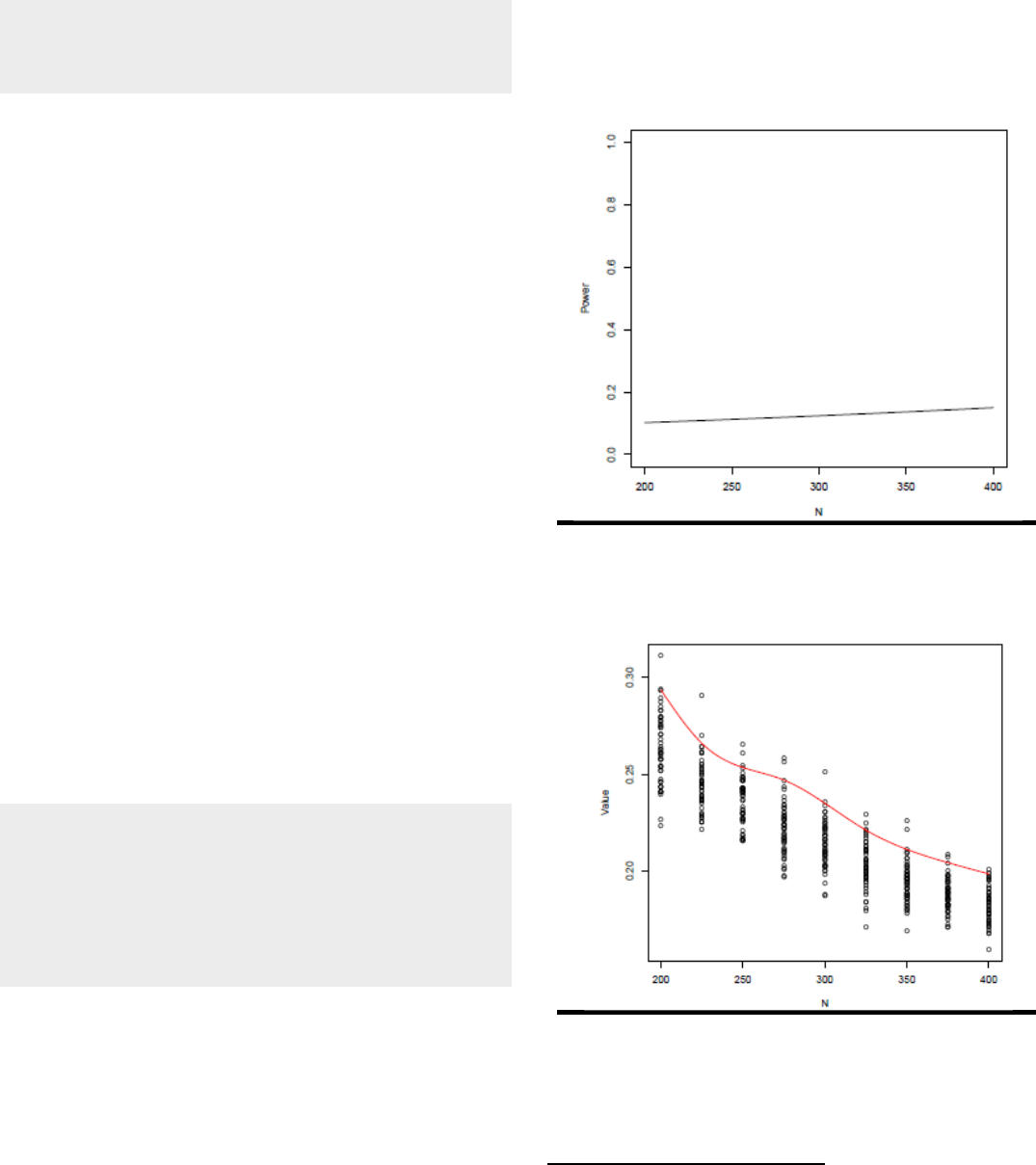
Power and accuracy
curves using sample sizes spanning 200-400 for the
1
2relation are shown in Figures 3a and 3b.
Figure 3a. Power curve for Example 1 using
α = .05.
Figure 3b. Accuracy curve for Example 1 using
α= .05.
2
I
ncorporating a level of
assurance
(i.e., probability) of the
CI’s half-width study is a
differen
t
form of the AIPE
perspective
than
I
discuss
in the
c
urren
t
article. For more information about
it,
see
Kelley and Maxwell (2008).

Practical Assessment, Research & Evaluation, Vol 19, No 12 Page 9
Beaujean, Monte Carlo Sample Size Determination
# power curve of the X2-Y relation
plotPower(analysis.n, powerParam = "y~x2", alpha
= 0.05)
# accuracy curve of the X2-Y relation
plotCIwidth(analysis.n, c("y~x2"), assurance = 0.95)
An alternative to graphically displaying the results
is to use the
getPower()
,
findPower()
, and
getCIwidth()
functions. The first and third functions return the
power and CI widths, respectively, for each parameter
at the specified sample sizes (
nVal
). The second
function uses a
getPower()
object to find the sample
size for a given level of power. If the
findPower()
function returns the values Inf or NA, it means the
sample size values are too large or too small,
respectively, for that parameter at the specified power
level.
# find n for power of .80
power.n <- getPower(analysis.n, alpha=.05,
nVal=200:300)
findPower(power.n, iv="N", power=0.80)
# find CI half-widths when n=200
getCIwidth(analysis.n, assurance = 0.95,
nVal=200)/2
Example 2: Multiple Regression
With Data Quirks
For this second example, I add some quirks to the
data from Example 2. I only present models that
include one quirk, but combining multiple quirks in a
single model is a simple extension.
Data Quirk 1: Missing Data
Missing data is often a problem in research, so
conducting a sample size analysis without accounting
for missing data is often unrealistic (Graham, 2009).
For the current example, I made 20% of X
2
's data
missing completely at random (MCAR), but X
1
missing
values dependent on values of X
3
. Specifically, when
X
3
= 0, 15% of X
1
's data is missing; for each unit
increase and decrease in X
3
, the amount of missing data
increases and decreases, respectively. As long as I
include X
3
in the regression model, the missing values
for X
1
are missing at random (MAR).
simsem
has a variety of ways to include missing
data in the simulations, most of which use the
miss()
function. I use the logit method because it has the
ability to graph the amount of missing data. The logit
method requires a lavaan-like script that specifies how
much data should be missing for a given variable. Each
line of the script begins with a variable, then the
regression symbol (
~
), and then values for the amount
of missing data.
The values after the
~
are input for the inverse
logit function:
345
.
6
7
8
1
7
8
1
7
9
/
7
345
.
6
7
8
1
7
8
1
7
9
/
(5)
where a is the intercept, the bs are slope values,
and the Xs are predictor variables in the regression. For
example, if a = –1.38 and there are no predictors, then
the inverse logit value is 0.20, so approximately 20% of
the values should be missing. Likewise, if a = –1.73 and
b = 0.25, then approximately 15% of the values are
missing when X = 0, 19% of the values when X = 1,
and so forth.
# specify amount of missing data using logit
method
pcnt.missing <- '
# 20% of data missing
x2 ~ -1.38
# 15% of X1 data is missing when X3 is zero
x1 ~ -1.73 + 0.25
*
x3
'
To plot the amount of missing data specified in
the logit equations, use the
plotLogitMiss()
function. The
plot for the current example is shown in Figure 4.
Figure 4 . Plot of missing data to include in
the simulated datasets for
1
and
1
.
# plot amount of missing data specified in the logit
equations
plotLogitMiss(pcnt.missing)

Practical Assessment, Research & Evaluation, Vol 19, No 12 Page 10
Beaujean, Monte Carlo Sample Size Determination
Having missing values requires determining how
to estimate the parameters in the presence of this
missingness. Only full information maximum
likelihood (FIML) and multiple imputation (MI) are
available estimation options in simsem, which can be
used with or without auxiliary variables. For details on
FIML, MI and auxiliary variables, see Enders (2011).
By default, simsem uses FIML when there are missing
values. To use MI requires two additional arguments:
the number of imputations for each data set (m) and
the R package to conduct the imputation (package)
3
.
Currently, only function from the mice (van Buuren &
Groothuis-Oudshoorn, 2011) and Amelia II (Honaker,
King, & Blackwell, 2011) packages can be used for the
imputation.
# FIML
missing.model.fiml <- miss(logit = pcnt.missing)
# MI
missing.model.mi <- miss(logit = pcnt.missing, m =
10, package = "mice")
# simulate regression data with missing data using
FIML
analysis.mis.237 <- sim(nRep=750,
model=analysis.model, n=237,
generate=pop.model, lavaanfun = "sem",
miss=missing.model.fiml, seed=565,
multicore=TRUE)
For the current example, I simulated the data with
n = 237 using FIML to handle the missing data. As I
included missing values, I increased m to 750. The
results are given in Table 4. The relative bias and
coverage values are within specified limits. Compared
to initial model (Example 1), power decreases slightly
for X
2
's and X
3
's regression coefficients and CI half-
widths for all thee predictors increase.
X
2
's regression coefficient is the smallest in value.
Thus, finding the sample size needed for it to be
estimated with power of 0.80, would mean that all
other regression coefficients would have at least a
power of 0.80. To find the sample size needed for X
2
's
regression coefficient to be estimated with power of
0.80, I use the same procedures described in finding an
unknown sample size for Example 1. As data are
missing, I specified the search to go from n = 200 to
3
The m
argumen
t
in the miss() function is not related to
th
e
number of simulated
samples
in the
MC
study, m.
Table 4.
Values
From Monte Carlo
Sample Size
Studies.
Relativ
e
Bias
95%
CI
Half-
Width
Pre-
dictor
Model
Value
Parameter
SE
Cover-
age
Power
No Data Quirks (Example
1)
X1
0.66
0.01
0.04
0.96
1.00
0.15
X2
0.05
-
0.09
0.00
0.95
0.12
0.12
X3
-
0.30
0.01
0.02
0.95
0.99
0.14
Missing
Data with Full Information Maximum Likelihood
Estimation
X1
0.66
-
0.00
0.03
0.96
1.00
0.18
X2
0.05
-
0.06
0.02
0.95
0.09
0.15
X3
-
0.30
0.01
0.01
0.95
0.94
0.17
Y
’s Distribution with
Excess Skew
and Kurtosis
X1
0.66
-
0.18
-
0.19
0.67
1.00
0.16
X2
0.05
-
0.15
-
0.02
0.95
0.12
0.13
X3
-
0.30
-
0.16
-
0.09
0.87
0.89
0.15
Unreliability of X
1
and
X
2
X1
0.66
0.12
-
0.03
0.99
0.48
0.90
X2
0.05
-
1.00
-
0.04
0.99
0.04
0.59
X3
-
0.30
0.16
-
0.03
0.97
0.25
0.54
n = 5000, increasing by 200, and using m = 50
simulations per sample size. The results indicate that
when n = 3820 power is .80. I already showed that with
n = 237 the CI half-with is 0.15; increasing it to 3820
makes the CI half-width approximately 0.06.
# sample size study for X2 using FIML to handle
missing data
# simulate data from n=200 to n=5000 by 25
missing.n <- sim(nRep=NULL,
model=analysis.model,
n=rep(seq(200,5000,200), 50),
generate=pop.model, lavaanfun = "sem",
miss=missing.model.fiml, multicore=TRUE)
# power curve
plotPower(missing.n, powerParam="y~x2",
alpha=.05)
# accuracy curve
plotCIwidth(missing.n, c("y~x2"), assurance = 0.95)
# find n for power of .80
power.mis <- getPower(missing.n, alpha=.05)
findPower(power.mis, iv="N", power=0.80)
# find CI half-widths
getCIwidth(missing.n, assurance = 0.95,
nVal=3820)/2
As a point of comparison, I examined the sample
size required for a power of .80 using traditional
methods. Specifically, I used the G*Power program

Practical Assessment, Research & Evaluation, Vol 19, No 12 Page 11
Beaujean, Monte Carlo Sample Size Determination
(Faul, Erdfelder, Buchner, & Lang, 2009) and followed
the steps the authors outlined for a power analysis of a
single regression coefficient in a multiple regression
(what they call Deviation of a Single Linear Regression
Coefficient From Zero). The values I used for the G*Power
program and its output are in Figure 5, which shows
that the sample size needed is 2057.
One way to handle missing data in this situation is
to divide the sample size required for a complete
dataset by the proportion of observations thought to be
without missing values. Thus, if 20% of the
observations had missing values, then divide n by .80 to
find the final sample size estimate. Assuming that
between 20-30% of the data are missing makes the
required sample size between 2571-2939, likely making
the study underpowered for X
2
's effects.
Data Quirk 2: Non-Normality
One assumption in multiple regression is that the
residuals are normally distributed (Williams, Grajales, &
Kurkiewicz, 2013). There are a variety of ways for the
residuals to fail to meet this assumption, but a common
one is for the outcome variable to have a non-normal
distribution, such as when it has excessive skew or
kurtosis. For the current example, I made Y's skew
equal to negative four and its kurtosis equal to seven. A
plot of such a variable is in Figure 6.
Figure 6 . Kernel density plot of Y with skew =
–4 and kurtosis = 7.
simsem’s
bindDist()
function makes any of the
variables in the simulated data have the desired amount
of skew and kurtosis using the
skewness
and
kurtosis
arguments, respectively. Skew and kurtosis values need
to be included for each variable in the data. By setting
the
indDist
argument equal to the
bindDist()
object, the
sim()
function uses the specified skew and kurtosis
values.
# add skew and kurtosis to only to Y
distrib <- bindDist(skewness = c(-4,0,0,0), kurtosis
= c(7,0,0,0))
Figure
5
. G*Power
value specification
for
example
with
missing
data.

Practical Assessment, Research & Evaluation, Vol 19, No 12 Page 12
Beaujean, Monte Carlo Sample Size Determination
# simulate data with non-normal Y
analysis.nn.237 <- sim(nRep=10000,
model=analysis.model, n=237,
generate=pop.model, lavaanfun = "sem",
indDist=distrib, seed=565, multicore=TRUE)
The results of the MC simulations with a non-
normal Y variable are in Table 4. With m = 750, the
relative parameter bias values are outside of the
specified limits for all three variables, as is the relative
SE bias and coverage for X
1
, and the coverage X
3
. The
aberrant bias values decrease minimally with a m of
10,000, indicating that using a typical regression model
for this data will produce biased regression coefficients.
Compared to the results from Example 1, the power
decreased for X
3
and the 95% CI half-widths increased
for all three predictors. As X
1
's relative SE bias is
somewhat large, its CI will likely be inaccurate so the
half-width should be interpreted cautiously.
Data Quirk 3: Reliability
Another assumption of multiple regression is that
the variables are measured without error. While ideal,
this is seldom the case for measures of psychological
constructs. Not accounting for measurement
unreliability in the model results in biased parameter
estimates and a decrease in power (Cole & Preacher,
2014).
One way to account for variables measured
without perfect reliability is to use single-indicator
latent variables (Keith, 2006). Single-indicator latent
variables explicitly model a variable’s variance, which is
what is affected with unreliable measures. If :
is a
variable’s variance and ;
<
is the reliability of a
variable’s scores, then single-indicator latent variables
fix the variable’s error variance to (1–;
<
) :
, the true
variance to (;
<
) :
, and the path coefficients to one.
To make single-indicator latent variables more
concrete, say the reliability of the scores for X
1
and X
2
are both .70. Figure 7 contains a path diagram of the
regression model with these imperfectly-measured
variables. Note the addition of the error and true score
components for X
1
and X
2
. In addition, while the
correlation and regression coefficients are the same as
those from Example 1, the residual variance has
increased from 0.685 to 0.816 because the R
2
decreased
after modeling their unreliability.
Figure 7 . Path model using single-indicator latent
variables to account for the imperfect reliability of
X
1
and X
2
, which is .70 for both variables. T
represents the true score and E represents
measurement error. The new R
2
value can be
calculated via:
.66 × .7 × .66 + .05 × .70 × .05 + –.30 × 1 × –.30
+ (.66 × .40 × .05) × 2 + (.66 × .60 ×
–.30) × 2 + (.05 × .05 × –.30) × 2 = .184.
# data generating model with measurement error
pop.rel.model <-'
# measurement model
x1.true =~ 1
*
x1
x2.true =~ 1
*
x2
# for reliabilities of .70
# constrain error variances of X1 and X2 to be .30
x1 ~~ .3
*
x1
x2 ~~ .3
*
x2
# constrain true score variances of X1 and X2 to be
.70
x1.true ~~ .7
*
x1.true
x2.true ~~ .7
*
x2.true
# regression
y ~ 0.66
*
x1.true + 0.05
*
x2.true + -0.30
*
x3
# predictor variables covariance
x1.true ~~ 0.40
*
x2.true + 0.60
*
x3
x2.true ~~ 0.05
*
x3
# residual variance
y~~ 0.816
*
y
'
# data analysis model accounting for measurement
error
analysis.rel.model <-'

Practical Assessment, Research & Evaluation, Vol 19, No 12 Page 13
Beaujean, Monte Carlo Sample Size Determination
# measurement model
x1.true =~ 1
*
x1
x2.true =~ 1
*
x2
# constrain error variances
x1 ~~ 0.3
*
x1
x2 ~~ 0.3
*
x2
# constrain true score variances
x1.true ~~ 0.7
*
x1.true
x2.true ~~ 0.7
*
x2.true
# predictor variables covariance
x1.true ~~ x2.true + x3 x2.true ~~ x3
# regression
y ~ x1.true + x2.true + x3
‘
# simulate data with unreliable variables
analysis.rel.237 <- sim(nRep = 750,
model=analysis.rel.model, n = 237,
generate=pop.rel.model, lavaanfun = "sem",
seed=565, multicore=TRUE
)
The results of the MC analysis are shown in Table
4. The relative parameter bias is a little high for X
1
and
X
3
, and very high for X
2
. When I set m = 5000, the
amount of relative parameter bias decreased to –.75 for
X
2
, indicating that even higher values of m might
produce better results for the MC study. The relative
SE bias and coverage are all within specified limits.
When compared to the values from Example 1, the
power for all three regression coefficients substantially
decreased and the 95% CI half-widths substantially
increased.
Discussion
In this article, I demonstrated the use of a Monte
Carlo (MC) study for the purpose of deciding on
sample size in regression models based on power and
accuracy in parameter estimation (AIPE). In the
examples, I used a multiple regression model with three
predictors and examined the sample size needed for
data without any quirks as well as data with missing
values, a non-normal outcome, and less-than-perfect
reliability. For the examples with no data quirks, the
results mapped directly onto the results from traditional
formula-based sample size determination methods.
When there are quirks in the data, however, there are
no simple formulae to determine sample size. The
results from the example MC studies showed that
ignoring these data quirks could result in underpowered
parameter estimation, inaccurate parameter estimates,
or both.
Unlike previous articles that showed how to use
MC studies to determine sample size, I focused on
regression models, since they are one of the most
common ways to analyze data (Troncoso Skidmore &
Thompson, 2010). In addition, I used the R statistical
language for all example analyses. As R is free and
available on many computer operating systems, the
procedures and R syntax in this article should be
readily usable by investigators for their own data
analysis.
Drawback of Using Monte Carlo Studies
While one of the purposes of this article was to
show the flexibility and benefits of the MC approach to
determining sample size, there is a drawback: it requires
users to know more about their studies’ variables than
traditional methods. Investigators have to specify not
onlyαand power, but they also have to specify values
for all the variables’ relations. Thus, Cohen’s (1992)
concern about researchers not knowing appropriate
effect size values for their particular field is amplified if
they have to know how all the variables relate to each
other. In the best situation, scholars would select the
model’s values from theory or previous research. In the
complete absence of any theoretical expectations,
Maxwell (2000) suggested starting with the assumption
that all zero-order correlations are .30, then changing
the values to see how it influences the required sample
size (i.e., sensitivity analysis). Using values of .30 gives
R
2
values around 0.14 (2 predictors) to 0.24 (10
predictors), which may or may not be appropriate for a
study.
Using the AIPE perspective adds yet one more
piece of information for the investigator to know: an
appropriate size for the CI half-width. There are
currently not any guidelines to determine the
appropriate CI half-width for a regression, but values
between 0.10 and 0.20 for standardized regression
coefficients are commonly used in the AIPE literature,
so are probably a good place to start. Of course, this
somewhat depends on the hypothesized value of the
regression coefficient. For example, narrower CIs are
likely better with coefficient values expected to be close
to zero in order to determine if the direction of the
effect is positive or negative. Likewise, if the coefficient
is expected to be large and well beyond some threshold
set for usefulness (e.g., a clinically-relevant effect), then

Practical Assessment, Research & Evaluation, Vol 19, No 12 Page 14
Beaujean, Monte Carlo Sample Size Determination
having a wider CI may be acceptable as long as its
bounds do not cross the threshold.
Monte Carlo Study Extensions
In the current article, I only focused on
determining the sample size for specific regression
coefficients. An alternative is to focus on the entire
model (i.e., omnibus) and base the sample size on the
R
2
value. This is easy to do using the simsem package
as the amount of error variance (i.e., 1–R
2
) is already
included as an estimated parameter in the output.
Another limitation of the models I used in this
article is that the outcome variable was continuous. The
same procedures could be used with variations of this
model, such as having categorical or count outcomes.
While this would require more complex models and
different effect sizes, the same basic procedures still
apply. Likewise, regression models with nested data
could also use this approach (e.g., Meuleman & Billiet,
2009).
References
American Educational Research Association. (2006).
Standards for reporting on empirical social science
research in AERA publications. Educational Researcher ,
35 , 33-40.
Bacchetti, P. (2010). Current sample size conventions:
Flaws, harms, and alternatives. BMC Medicine, 8 , 17.
Bacchetti, P. (2013). Small sample size is not the real
problem. Nature Reviews Neuroscience, 14 , 585-585. doi:
10.1038/nrn3475-c3
Barrett, P. (2007). Structural equation modelling: Adjudging
model fit. Personality and Individual Differences, 42 , 815-
824. doi: 10.1016/j.paid.2006.09.018
Beasley, W. H., & Rodgers, J. L. (2012). Bootstrapping and
Monte Carlo methods. In H. Cooper, P. M. Camic, D.
L. Long, A. T. Panter, D. Rindskopf, & K. J. Sher
(Eds.), APA handbook of research methods in psychology, Vol
2: Research designs: Quantitative, qualitative, neuropsychological,
and biological (pp. 407–425). Washington, DC: American
Psychological Association.
Beaujean, A. A. (2014). Latent variable modeling using
R
: A step-
by-step guide. New York, NY: Routledge/Taylor and
Francis.
American Psychological Association Publications and
Communications Board Working Group on Journal
Article Reporting Standards. (2008). Reporting
standards for research in psychology: Why do we need
them? What might they be? American Psychologist, 63 ,
839-851. doi: 10.1037/0003-066X.63.9.839
R Development Core Team. (2014).
R
: A language and
environment for statistical computing [Computer program].
Vienna, Austria: R Foundation for Statistical
Computing.
Boker, S. M., & McArdle, J. J. (2005). Path analysis and path
diagrams. In B. Everitt & D. C. Howell (Eds.),
Encyclopedia of behavioral statistics (Vol. 3, pp. 1529–1531).
West Sussex, England: Wiley.
Christensen, R. (2002). Plane answers to complex questions: The
theory of linear models (3rd ed.). New York, NY: Springer-
Verlag.
Cohen, J. (1988). Statistical power analysis for the behavioral
sciences (2nd ed.). Mahwah, NJ: Erlbaum.
Cohen, J. (1992). A power primer. Psychological Bulletin, 112 ,
155-159. doi: 10.1037/0033-2909.112.1.155
Cole, D. A., & Preacher, K. J. (2014). Manifest variable path
analysis: Potentially serious and misleading
consequences due to uncorrected measurement error.
Psychological Methods, 19, 300-315. doi:
10.1037/a0033805
Cumming, G. (2014). The new statistics: Why and how.
Psychological Science, 25 , 7-29. doi:
10.1177/0956797613504966
Cumming, G., & Finch, S. (2005). Inference by eye:
Confidence intervals and how to read pictures of data.
American Psychologist, 60 , 170-180. doi: 10.1037/0003-
066X.60.2.170
Descôteaux, J. (2007). Statistical power: An historical
introduction. Tutorials in Quantitative Methods for
Psychology, 3 (2), 28-34. Retrieved from
http://www.tqmp.org/Content/vol03-
2/p028/p028.pdf
Dupont, W. D., & Plummer, W. D., Jr.. (1998). Power and
sample size calculations for studies involving linear
regression. Controlled Clinical Trials, 19 , 589-601. doi:
10.1016/S0197-2456(98)00037-3
Enders, C. K. (2011). Applied missing data analysis. New York,
NY: Guilford.
Fan, X. (2012). Designing simulation studies. In H. Cooper
(Ed.), APA handbook of research methods in psychology (Vol.
3.): Data analysis and research publication (p. 427-444).
Washington, DC: American Psychological Association.
Faul, F., Erdfelder, E., Buchner, A., & Lang, A.-G. (2009).
Statistical power analyses using G*Power 3.1: Tests for
correlation and regression analyses. Behavior Research
Methods, 41 , 1149-1160. doi: 10.3758/brm.41.4.1149
Faul, F., Erdfelder, E., Lang, A.-G., & Buchner, A. (2007).
G*Power 3: A flexible statistical power analysis

Practical Assessment, Research & Evaluation, Vol 19, No 12 Page 15
Beaujean, Monte Carlo Sample Size Determination
program for the social, behavioral, and biomedical
sciences. Behavior Research Methods, 39 , 175-191. doi:
10.3758/bf03193146
Goodman, S. N., & Berlin, J. A. (1994). The use of predicted
confidence Intervals when planning experiments and
the misuse of power when interpreting results. Annals of
Internal Medicine, 121 , 200-206. doi: 10.7326/0003-
4819-121-3-199408010-00008
Graham, J. W. (2009). Missing data analysis: Making it work
in the real world. Annual Review of Psychology, 60 , 549-
576. doi: 10.1146/annurev.psych.58.110405.085530
Grissom, R. J., & Kim, J. J. (2005). Effect sizes for research: A
broad practical approach. Mahwah, NJ: Lawrence Erlbaum.
Hellmann, J. J., & Fowler, G. W. (1999). Bias, precision, and
accuracy of four measures of species richness. Ecological
Applications, 9 , 824-834. doi: 10.1890/1051-
0761(1999)009[0824:BPAAOF]2.0.CO;2
Hill, C. J., Bloom, H. S., Black, A. R., & Lipsey, M. W.
(2008). Empirical benchmarks for interpreting effect
sizes in research. Child Development Perspectives, 2 , 172-
177. doi: 10.1111/j.1750-8606.2008.00061.x
Honaker, J., King, G., & Blackwell, M. (2011). Amelia II: A
program for missing data. Journal of Statistical Software,
45(7), 1-47. Retrieved from
http://www.jstatsoft.org/v45/i07
Hoyle, R. H., & Smith, G. T. (1994). Formulating clinical
research hypotheses as structural equation models: A
conceptual overview. Journal of Consulting and Clinical
Psychology, 62 , 429-440. doi: 10.1037/0022-
006X.62.3.429
Keith, T. Z. (2006). Multiple regression and beyond. Boston:
Pearson.
Kelley, K. (2007). Methods for the Behavioral, Educational,
and Social Sciences: An R package. Behavior Research
Methods, 39 , 979-984. doi: 10.3758/BF03192993
Kelley, K., Lai, K., & Wu, P.-J. (2008). Using R for data
analysis: A best practice for research. In J. W. Osborne
(Ed.), Best practices in quantitative methods (pp. 535-572).
Thousand Oaks, CA: Sage.
Kelley, K., & Maxwell, S. E. (2003). Sample size for multiple
regression: Obtaining regression coefficients that are
accurate, not simply significant. Psychological Methods, 8 ,
305-321. doi: 10.1037/1082-989X.8.3.305
Kelley, K., & Maxwell, S. E. (2008). Sample size planning
with applications to multiple regression: Power and
accuracy for omnibus and targeted effects. In P.
Alasuutari, L. B. Bickman, & J. Brannen (Eds.), The Sage
handbook of social research methods (pp. 166-192).
Thousand Oaks, CA: Sage.
Kelley, K., & Maxwell, S. E. (2012). Sample size planning. In
H. Cooper, P. M. Camic, D. L. Long, A. T. Panter, D.
Rindskopf, & K. J. Sher (Eds.), APA handbook of research
methods in psychology, Vol 1: Foundations, planning, measures,
and psychometrics (pp. 181–202). Washington, DC:
American Psychological Association.
Marsaglia, G. (2003). Seeds for random number generators.
Communications of the ACM , 46 , 90-93. doi:
10.1145/769800.769827
Maxwell, S. E. (2000). Sample size and multiple regression
analysis. Psychological Methods, 5 , 434-458. doi:
10.1037/1082-989X.5.4.434
Maxwell, S. E., Kelley, K., & Rausch, J. R. (2007). Sample
size planning for statistical power and accuracy in
parameter estimation. Annual Review of Psychology, 59 ,
537-563. doi:
10.1146/annurev.psych.59.103006.093735
Meuleman, B., & Billiet, J. (2009). A Monte Carlo sample
size study: How many countries are needed for accurate
multilevel SEM? Survey Research Methods, 3 (1), 45-58.
Retrieved from http://ojs.ub.uni-
konstanz.de/srm/article/view/666.
Moher, D., Hopewell, S., Schulz, K. F., Montori, V.,
Gøtzsche, P. C., Devereaux, P. J., Altman, D. G.
(2010). CONSORT 2010 Explanation and Elaboration:
updated guidelines for reporting parallel group
randomised trials. Journal of Clinical Epidemiology, 63 , e1-
e37. doi: 10.1016/j.jclinepi.2010.03.004
Murphy, K. R., & Myors, B. (1998). Statistical power analysis: A
simple and general model for traditional and modern hypothesis
tests. Mahwah, NJ: Erlbaum.
Muthén, L. K., & Muthén, B. O. (2002). How to use a
Monte Carlo study to decide on sample size and
determine power. Structural Equation Modeling: A
Multidisciplinary Journal , 9 , 599-620. doi:
0.1207/S15328007SEM0904_8
Pornprasertmanit, S., Miller, P., & Schoemann, A. (2012).
simsem: SIMulated Structural Equation Modeling [Computer
software]. Retrieved from http://CRAN.R-
project.org/package=simsem
Rosseel, Y. (2012). lavaan: An R package for structural
equation modeling. Journal of Statistical Software, 48 (2), 1-
36. Retrieved from http://www.jstatsoft.org/v48/i02
Skrondal, A. (2000). Design and analysis of Monte Carlo
experiments: Attacking the conventional wisdom.
Multivariate Behavioral Research, 35 , 137-167. doi:
10.1207/S15327906MBR3502_1
Troncoso Skidmore, S., & Thompson, B. (2010). Statistical
techniques used in published articles: A historical
review of reviews. Educational and Psychological

Practical Assessment, Research & Evaluation, Vol 19, No 12 Page 16
Beaujean, Monte Carlo Sample Size Determination
Measurement, 70 , 777-795. doi:
10.1177/0013164410379320
van Buuren, S., & Groothuis-Oudshoorn, K. (2011).
Multivariate imputation by chained equations in R.
Journal of Statistical Software, 45 (3). Retrieved from
http://www.jstatsoft.org/v45/i03
Venables, W. N., Smith, D. M., & R Development Core
Team. (2012). An introduction to R. R Development
Core Team. Retrieved from http://cran.r-
project.org/doc/manuals/R-intro.pdf
Wilkinson, L., & American Psychological Association
Science Directorate Task Force on Statistical Inference.
(1999). Statistical methods in psychology journals:
Guidelines and explanations. American Psychologist, 54 ,
594-604. doi:10.1037/0003-066X.54.8.594
Williams, M. N., Grajales, C. A. G., & Kurkiewicz, D.
(2013). Assumptions of multiple regression: Correcting
two misconceptions. Practical Assessment Research and
Evaluation, 18 (11), 1-14. Retrieved from
http://pareonline.net/getvn.asp?v=18&n=11
Citation:
Beaujean, A. Alexander (2014). Sample Size Determination for Regression Models Using Monte Carlo
Methods in R. Practical Assessment, Research & Evaluation, 19(12). Available online:
http://pareonline.net/getvn.asp?v=19&n=12
Author:
A. Alexander Beaujean, Director
Baylor Psychometric Laboratory
Department of Educational Psychology
One Bear Place #97301
Waco, TX 76798-7301
Alex_Beaujean [at] Baylor.edu
http://blogs.baylor.edu/alex_beaujean
